NMR-techniques in Drug Research
On behalf of the course team, I would like to welcome to the 2020 edition of our PhD course NMR Techniques in Drug Research- NTDR2020. We hope that the course will provide you with a stimulating environment for learning and exploring the possible uses of NMR spectroscopy in drug research, and will result in a more extensive use of NMR methods in your own research
The course team consist of five teachers - all using NMR spectroscopy in various aspects of drug research. We are confident that we can provide you with inspiration and knowledge about advanced NMR spectroscopic techniques used in drug research. The course team consist of:
- Arife Önder, Laboratory technician (Department of Drug design and Pharmacology)
- Chao Liang, Postdoc (Department of Drug design and Pharmacology)
- Dan Stærk, Professor (Department of Drug Design and Pharmacology)
- Kenneth T. Kongstad, Assistant Professor (Department of Drug Design and Pharmacology)
- Louise Kjærulff, Postdoc (Department of Drug design and Pharmacology
An overview of the course content can be seen in the attached file ‘NTDR2020 Course programme overview’ and ‘NTDR2020 Course programme’. During the course, you will have hands-on experience with NMR spectrometers. Thus, the course is composed of lectures as well as practical sessions comprising sample preparation, acquisition of NMR data, and computer exercises (data processing and interpretation):
- Spectrometer optimization (determination of 90° pulses, probe tuning and matching)
- Determination of longitudinal relaxation times (T1)
- Structure determination by 2D NMR (pre-acquired dataset)
- Peptide NMR (pre-acquired dataset)
- STD NMR – exploring ligand-enzyme interactions by saturation transfer NMR
- Structure determination by 2D NMR (students prepare sample & acquire dataset)
- Determination of absolute configuration by the Mosher method
- Metabolomics
- Computer-aided structure elucidation
All participants must prepare short written reports about the exercises performed. The reports should be prepared and submitted during the course. There is a limited possibility to carry out exercises with problems suggested by the participants. If you are interested in this, please contact Professor Dan Stærk as soon as possible.
We have a very busy schedule during the 8 course days - and thus it is advantageous to do some background reading before course start. I have attached background reading for the first four lectures which I would like you to read before March 23rd. Notice that you should be selective in your reading!! Thus, book chapters should be read more thoroughly than experimental details in the scientific papers. I trust that you as responsible and experienced PhD students will be able to make proper decisions on how extensive the different topics should be read. The remaining background reading will become available at the course home page very soon.
Evaluation (passed/not passed) will be based on active participation, the above reports and a short 15 min presentation of results obtained during the course.
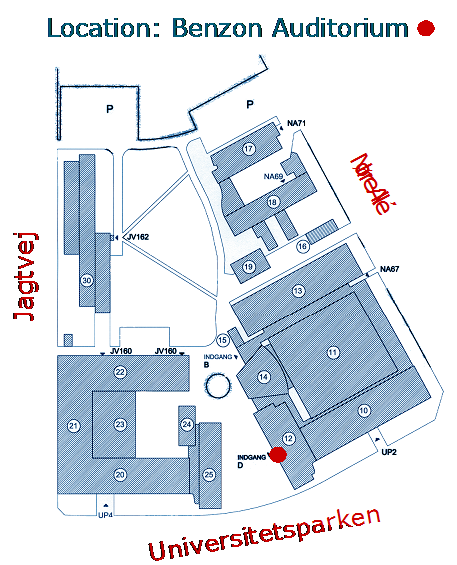
The course takes place in room U22 at Jagtvej 160, 2100 Copenhagen (Østerbro). Please use the entrance to building 22,go up the stairs and take either the stairs or lift to the 4th level (see map).
Lectures will be held in U22, and laboratory exercises will take place in either U22 or in the NMR laboratory in building 30.
Breakfasts will be served outside U22, whereas combined group discussions/lunch will take place in the student cafeteria in building 23.
We will use Topspin 4.0.8 for data processing during the course. A free version can be installed on your Microsoft Windows laptop or you MacBook. Go to the following url: https://www.bruker.com/service/support-upgrades/software-downloads/nmr.html and log in (upper right corner) by setting up an account (any mail will do + password). This will bring you back to the same page but now as a registered user. Download 'the latest version' of Topspin, then go to the page: https://www.bruker.com/service/support-upgrades/software-downloads/nmr/free-topspin-processing/topspin-demo-licences-generation.html where you can request a license code for the free demo license. If you have already installed Topspin 4 with the academic free version, you should also request for the free demo license and then go to http://licensecentral-bbio.bruker.de to activate the demo license too, so you can use CMC-se. Please check whether the CMC-se add-on is working in Topspin by issuing the command “cmcse” in the status line in the lower left corner of the Topspin 4 window. If “Project management” pops up, it works!
Course team members
| Name | Title | |
|---|---|---|
| Kjærulff, Louise | Research Consultant |
|