Computational Receptor Biology
– Gloriam Group
David Gloriam’s group identifies mechanisms and ligands of G protein-coupled receptors (GPCRs) – the main mediators of human signalling and drug responses – and develop the field’s main online database, GPCRdb (see video and GPCRdb.org).
Follow us
Google Scholar profile
Scopus publication list
GPCR drug discovery: new agents, targets and indications
Lorente JS, Sokolov AV, Ferguson G, Schiöth HB, Hauser AS, Gloriam DE
Nature Reviews Drug Discovery 2025
An online GPCR drug discovery resource
Caroli J, Andreassen SN, Lorente JS, Xiao B, Pándy-Szekeres G, Gloriam DE
npj Drug Discovery 2025
Structural basis of Δ9-THC analog activity at the Cannabinoid 1 receptor.
Thorsen T, Kulkarni Y, Sykes D, Bøggild A, Drace T, Hompluem P, Iliopoulos-Tsoutsouvas C, Nikas S, Daver H, Makriyannis A, Nissen P, Gajhede M, Veprintsev D, Boesen T, Kastrup J, Gloriam D
Nature Communications 2025
GPCRdb in 2025: adding odorant receptors, data mapper, structure similarity search and models of physiological ligand complexes. Herrera LPT, Andreassen SN, Caroli J, Rodríguez-Espigares I, Kermani AA, Keserű GM, Kooistra AJ, Pándy-Szekeres G, Gloriam DE.
Nucleic Acids Research 2025
Functional dynamics of G protein-coupled receptors reveal new routes for drug discovery.
Conflitti P, Lyman E, Sansom MSP, Hildebrand PW, Gutiérrez-de-Terán H, Carloni P, Ansell TB, Yuan S, Barth P, Robinson AS, Tate CG, Gloriam D, Grzesiek S, Eddy MT, Prosser S, Limongelli V.
Nature Reviews Drug Discovery 2025
GproteinDb in 2024: new G protein-GPCR couplings, AlphaFold2-multimer models and interface interactions.
Pándy-Szekeres G, Taracena Herrera LP, Caroli J, Kermani AA, Kulkarni Y, Keserű GM, Gloriam DE
Nucleic Acids Research 2024
A community Biased Signaling Atlas
Caroli J, Mamyrbekov A, Harpsøe K, Gardizi S, Dörries L, Ghosh E, Hauser AS, Kooistra AJ, Gloriam DE
Nature Chemical Biology 2023
Structural insights into the lipid and ligand regulation of serotonin receptors
Xu P, Huang S, Zhang H, Mao C, Zhou E, Cheng X, Simon IA, Shen DD, Yen HS, Robinson CV, Harpsøe K, Svensson B, Jiang H, Gloriam DE, Melcher K, Jiang Y, Zhang Y and Xu HE
Nature 2021
Discovery of human signaling systems - pairing peptides to G protein-coupled receptors
Foster SR, Hauser AS, Vedel L, Strachan RT, Huang XP, Gavin AC, Shah SD, Nayak AP, Haugaard-Kedström LM, Penn RB, Roth BL,Bräuner-Osborne H* and Gloriam DE*
Cell.2019
Current funding


Previous funding





Jody Pacalon wins best poster prize
2025.07.09
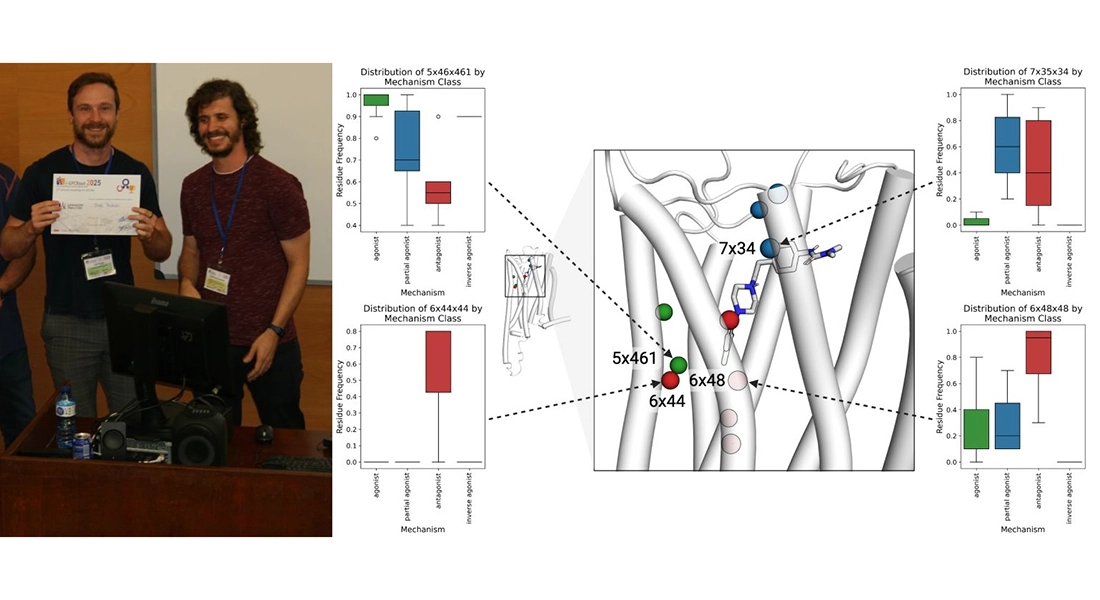 Jody Pacalon received the best postdoc poster prize at the 5th Annual iGPCRnet Meeting in Barcelona. Jody combines molecular dynamics simulations with in vitro pharmacological data to identify key interaction hotspots at the amino acid level.
Jody Pacalon received the best postdoc poster prize at the 5th Annual iGPCRnet Meeting in Barcelona. Jody combines molecular dynamics simulations with in vitro pharmacological data to identify key interaction hotspots at the amino acid level.
These insights help guide the rational design of ligands with improved selectivity and functional profiles better antipsychotic treatments.
Silver in group modeling competition
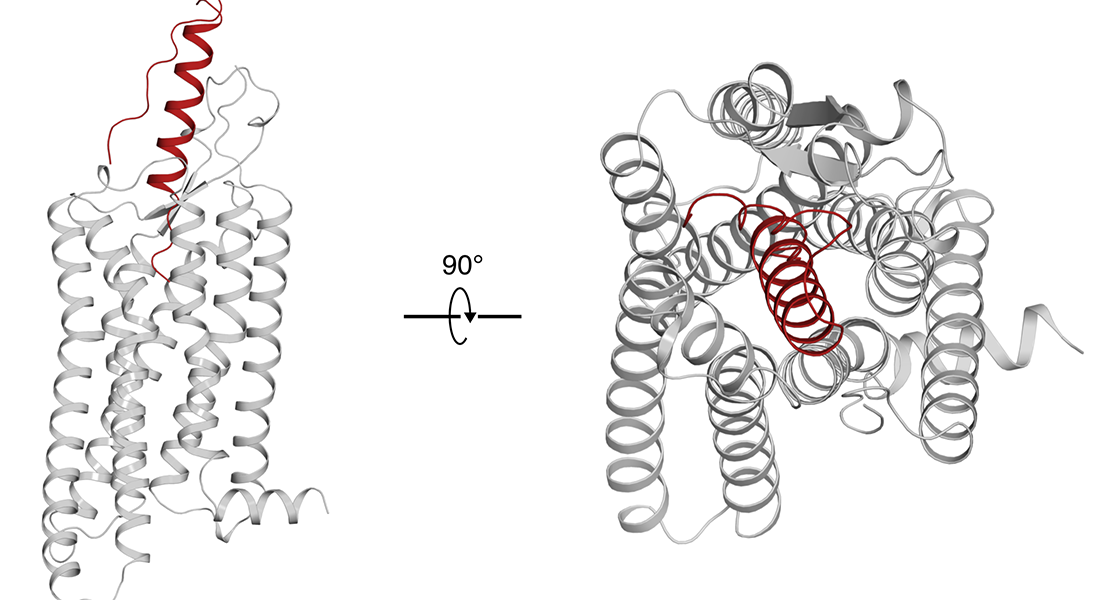
The Gloriam group achieved the second-best scoring model of the neuropeptide Y – NPY1 complex in the 4th GPCR Dock – a global competition assessing structure/ligand modeling for GPCR targets. Furthermore, it got the best pocket RMSD for the apelin receptor (APJ21) and second-best ligand (dynorphin) RMSD for the k-opioid receptor (8F7W_core). See pre-print.
Albert J. Kooistra receives the MGMS Frank Blaney Award
2021.11.27

David Gloriam receives big data prize from the Danish Royal Academy of Sciences and Letters
2021.10.08

Alexander S. Hauser receives Bachem Award for Peptide Science
2021.03.26

Alexander Hauser receives HC Ørsted research talent prize
2019.08.14

Professor inauguration
09.05.2019
Inauguration of David Gloriam as Professor. … »

Crystal refinement bronze
15.11.2016
Bronze prize in crystal structure refinement.
Louise Nikolajsen got
Wikipedia gold
11.07.2016
Gold prize in Wikipedia competition.
The International Society for Computational Biology (ISCB) announces competitions to improve the coverage on Wikipedia of any aspect of computational biology. In the last WikiProject competition PhD student Alexander S. Hauser has been awarded 1st and 2nd prize awards with an in-depth update of the molecular docking Wikipedia page as well as Wikidata entry.

Oral & poster awards
01.06.2016
2nd Central European Biomedical Congress.
Stefan Mordalski and Gáspár Pándy-Szekeres received awards for their oral and poster presentations, respectively, at the 2nd Central European Biomedical Congress, in Krakow, Poland.

GPCR modelling gold
01.08.2014
Best 5-HT1B model in global competition.
1st and 3rd places in the GPCR Dock competition for structure
2025 03 25
Mechanisms of THC and potential for designing medicines
2025 03 04
Major analysis identifies potential new drug targets
GPCRdb accelerates data-driven receptor research
2021 01 18
GPCRdb accelerates data-driven receptor research
Discovery of new hormone-receptor signalling systems
2019.10.31
Press release and video
Ultra-large virtual screening for drugs
2019.02.06
News & views article and podcast
Press release: GPCR structure determination resource
2019.01.23
Online tools to determine GPCR 3D structures.
The first web-based tool to determine GPCR 3D structures (Nature Methods) - Eng
Press release: 5-HT2C structure
2018.02.01
Shedding insight on GPCR-ligand polypharmacology.
Polypharmacology and 3D structure of the serotonin 5-HT2C receptor (Cell) - Eng
Video on GPCR pharmacogenomics
2018.01.11
From the Cell article on pharmacogenomics of GPCR drug targets. … »
Press release: GPCR pharmacogenomics
2017.12.15
Genetic variation in GPCR drug targets.
Pharmacogenomics of GPCR drug targets (Cell) - Eng1, Eng2 & Dan
Press release: GPCR drug discovery
2017.10.31
Trends for drugs, clinical trials and indications.
Trends in GPCR drug discovery (Nat Rev Drug Discov) - Eng & Dan
Press release: G protein selectivity
2017.05.11
Determinants of GPCR-G protein coupling.
Receptor signalling: GPCR - G protein selectivity (Nature) - Eng & Dan
Intro to the GPCRdb database
2015.06.01
Popular scientific article about the GPCRdb database.
Video on orphan receptor ligand identification
2015.01.01
Video made in relation to the award of the Lundbeck Foundation Fellowship to David Gloriam. English: … »
Research
We combine bioinformatics, chemoinformatics and computational chemistry in computational drug design at GPCRs. We also develop new methods for virtual screening, chemogenomics and design of target-directed focused screening libraries.
The work has led to the identification of novel ligands for the chemokine, glutamate, histamine and serotonin receptors. It is highly interdisciplinary, building on collaborations with Medicinal Chemistry and Molecular Pharmacology groups at the Department of Drug Design and Pharmacology, as well as several international labs.
The explosion of biomedical data in genomics, structural biology and pharmacology provides new opportunities to deepen our understanding of human physiology and disease. We integrate these data with innovative computational tools to gain novel insights into receptor biology. Our strength is the combined expertise in selected biological systems with the integration of diverse often unique datasets and hypotheses. The common objective of our projects is to tackle some of the currently most sought-after questions in the GPCR field.

GPCRdb is a major hub for the GPCR field and serves 50,000 users every year. GproteinDb, ArrestinDb and Biased Signaling Atlas cover related aspects of signal transduction while serving dedicated research communities. These online research infrastructures serve science through reference data, analysis, visualization, experiment design and data deposition.
Group members
| Name | Title | |
|---|---|---|
| Andreassen, Søren Norge | Academic Research Staff |
|
| Durdevic, Anja | Master Thesis Student |
|
| Henseler, Jascha | PhD Fellow |
|
| Kulkarni, Yashraj | Postdoc |
|
| Pándy-Szekeres, Gáspár | Academic Research Staff |
|
| Simonyan, Arman | PhD Fellow |
|
| Sánchez Lorente, Javier | PhD Fellow |
|
| Xiao, Binghan | PhD Fellow |
|
Persons are listed with year of leaving and next job
Postdoc & Assistant Professors
Albert Kooistra, 2022, Associate Professor at UCPH
Alexander Hauser, 2022, Associate Professor at UCPH
Mauricio Esguerra, 2021, Nostrum Bioscience
Thor Thorsen, 2021, Ebumab ApS
Luis Taracena, 2024
Kay Schaller, 2023, Novo Nordisk A/S
Eshan Ghosh, 2022, AstraZeneca
Henrik Daver, 2021, Lundbeck A/S
Stefan Mordalski, 2020, Intomics A/S
Louise F. Nikolajsen, 2019, Novo Nordisk A/S
Vignir Isberg, 2016, Novozymes A/S
Kasper Harpsøe, 2015, Facilitaty manager at UCPH
PhD students
Alibek Mamyrbekov, 2021, Novo Nordisk A/S
Cecilie Bonnesen, 2021, Fujifilm Danmark A/S
Icaro Simon, 2021, Vrije Universiteit Amsterdam
Mads Nygaard, 2021, Gubra
Christian Munk, 2020, Novozymes A/S
Alexander Hauser, 2019
Tsonko Tsonkov, 2019, Novo Nordisk A/S
Louise F. Nikolajsen, 2018, Novo Nordisk A/S
Mohamed Shehata, 2018, Novo Nordisk A/S
Vignir Isberg, 2014
Research assistants
Christian Breinholt, 2021, Semler gruppen A/S
Sahar Gardizi, 2020, Novo Nordisk A/S
MSc thesis project
Oulin Zhang, 2025
Felix Weckerle, 2024
Jakob Sture Madsen, 2021, Octant
Stylianos Iliadis, 2021, PhD studies at Queen Mary University of London
Alicia Bergkvans, 2018, PhD studies at VU Amsterdam
Palash Jain, 2018, University of Nottingham
Alexander Nielsen, 2017, Novo Nordisk A/S
Anders Nielsen, 2017
Anna Tsolakou, 2017
Dagmar A. Dalin, 2017
Leonard Floryan, 2017
Nadia Lund, 2017
Szonja Fehertavi, 2017
Christian Munk, 2016
Alexander Hauser, 2015
Gaspar Pandy-Szekeres, 2015
Kimberley Fidom, 2014
Mohamed Shehata, 2014
Alexander Chalikiopoulos, 2013
Kang Li, 2013
Thomas Lehto, 2013
Theo Jepsen, 2013
Magnus_Vesterager_Larsen, 2012
Andrius Senulis, 2012
Rasmus Leth, 2012
Vignir Isberg, 2010
Guest researchers
Janne Bibbe, 2017, PhD studies at Utrecht University
Willem Jespers, 2016, Assistant Professorship at University of Groningen
Eshita Mutt, 2015
Juan Carlos Mobarec, 2014, Heptares
Valentina Sepe, 2014, Associate Professor at University of Naples

