Pharmacoinformatics – Hauser Group
We integrate large biomedical data in genomics, structural biology, pharmacology, and health care combining innovative computational methods to gain insights into novel drug targets and to reveal fundamental principles in biological systems.
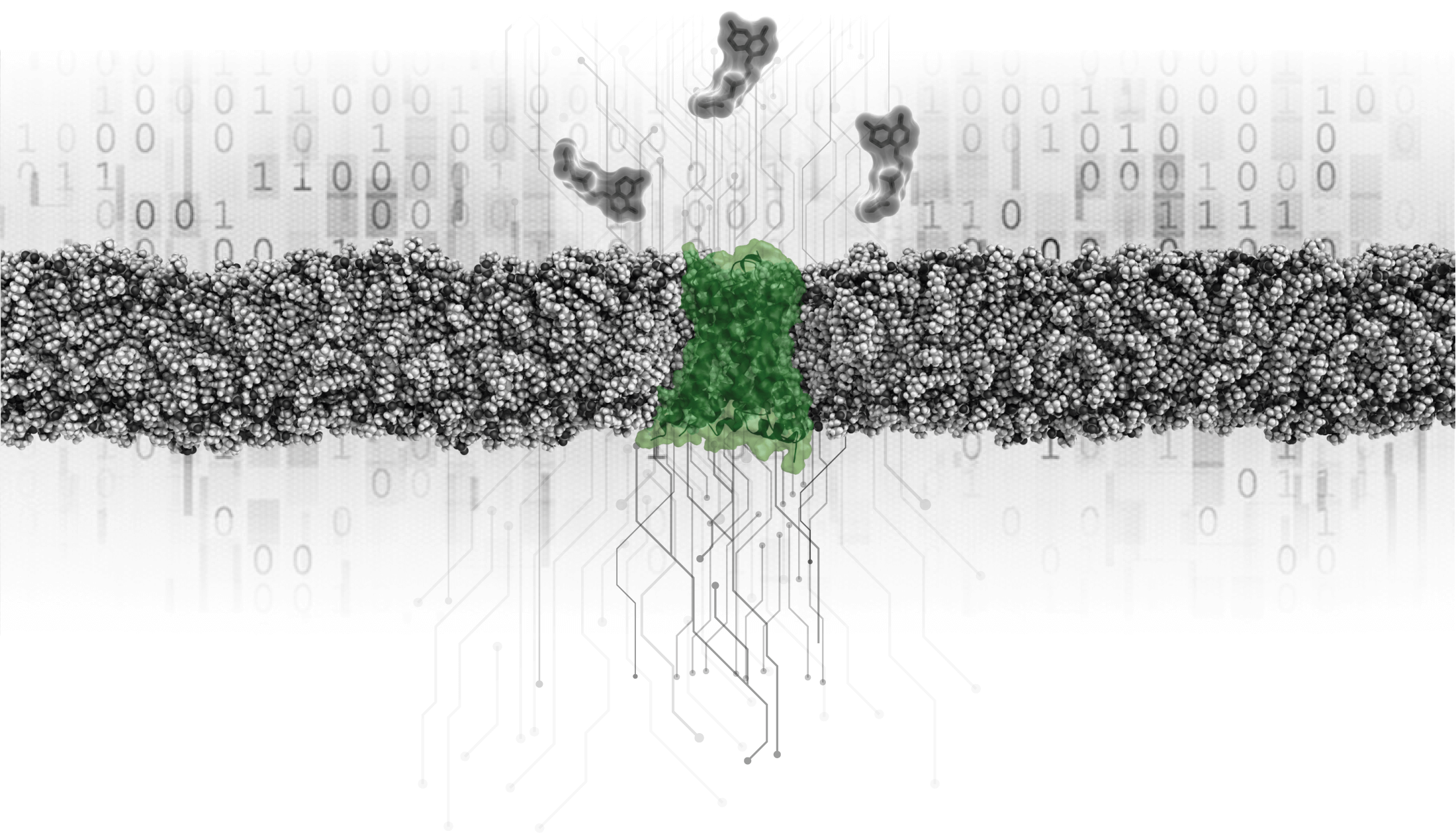
Research
The explosion of biomedical data in genomics, structural biology, pharmacology, and related fields provide new opportunities to deepen our understanding of human physiology and disease. We integrate these data with innovative computational tools to gain novel insights into protein biology.
Our strength is the combined expertise in selected biological systems with the integration of diverse often unique datasets and hypotheses. The common objective of our projects is to tackle some of the currently most sought-after questions in the GPCR field.
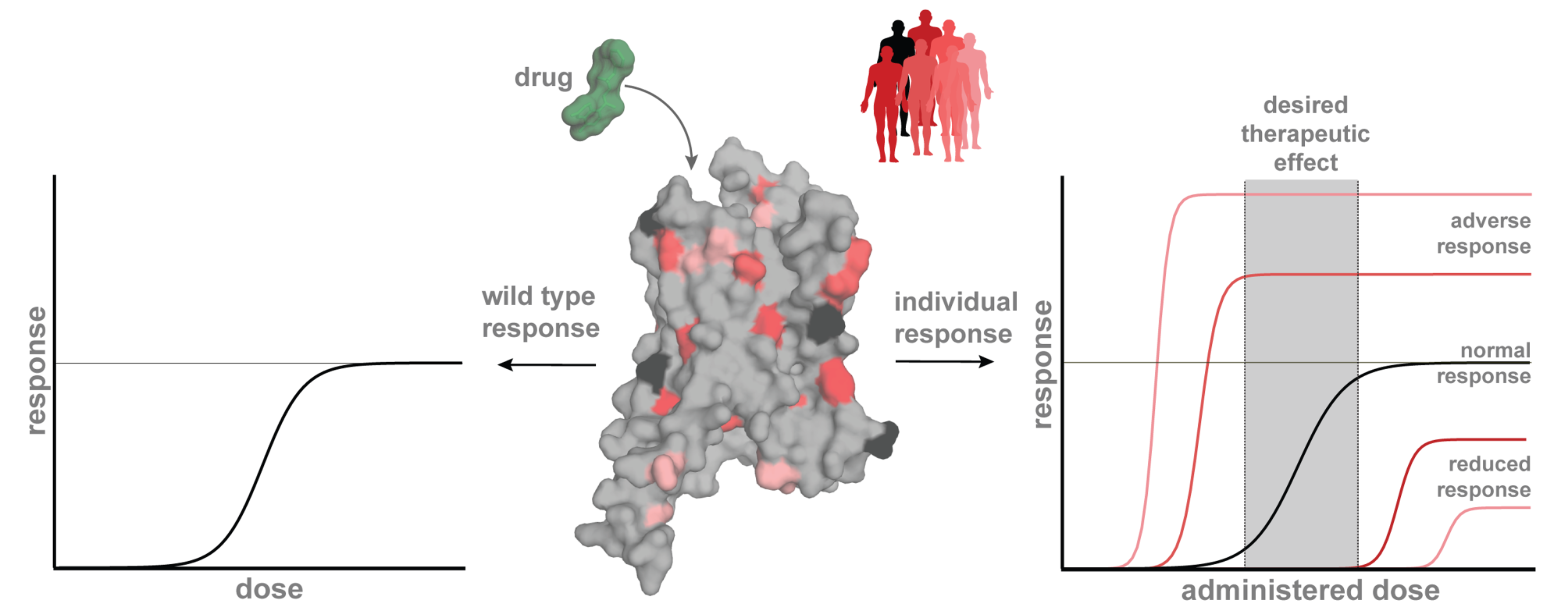
The individual phenotypic and molecular response to a given therapeutic treatment is a convoluted trail, shaped by inherited and environmental factors. The genetic variation across individuals leads to personal differences in the metabolism, therapeutic effects and adverse reactions of drugs.
Pharmacogenomics aims to target treatments by considering each individual’s genetic makeup. The main goal of personalised medicine is to tailor drug prescriptions to individuals to achieve the most effective and least harmful treatment. This is important to quicker find the optimal of multiple drugs and to avoid unnecessarily high dosages.
Key to realising the potential of personalised medicine is integrating systems biology, statistics and bioinformatics to associate genotypes to phenotypes. Our work bridges the fields of genetics and genomics, structural biology, pharmacology, drug development and medicine, as a case study for GPCRs.
This is very timely considering the novel big datasets which combined with high-throughput pharmacology, international biobanks such as the UK Biobank, and the unique Danish registries provide an innovative new approach to advance personalised medicine.
On this foundation, we are interested in:
1. Predicting how mutations can affect drug response (in silico)
2. Determine how drug responses are affected by genetic variations (in vitro)
3. Delineate genetic variants associated with physiology and disease or a change of clinical therapy (in human)
4. Create a public platform to phenotype GPCR variants for personalised medicine (in field)
Our linking of missense variants to patient disease cohorts will be very valuable for:
1: more accurate patient sub-diagnoses for genotype-based personalised treatment.
2: patient stratification upon entering clinical trials, and
3: increased precision in delineation of changes in clinical therapy,
4: personalise medicine prescriptions based on genotypes,
5: prioritise drugs for pharmacovigilance investigations, and
6: design post-market follow-up studies e.g. drug repurposing.
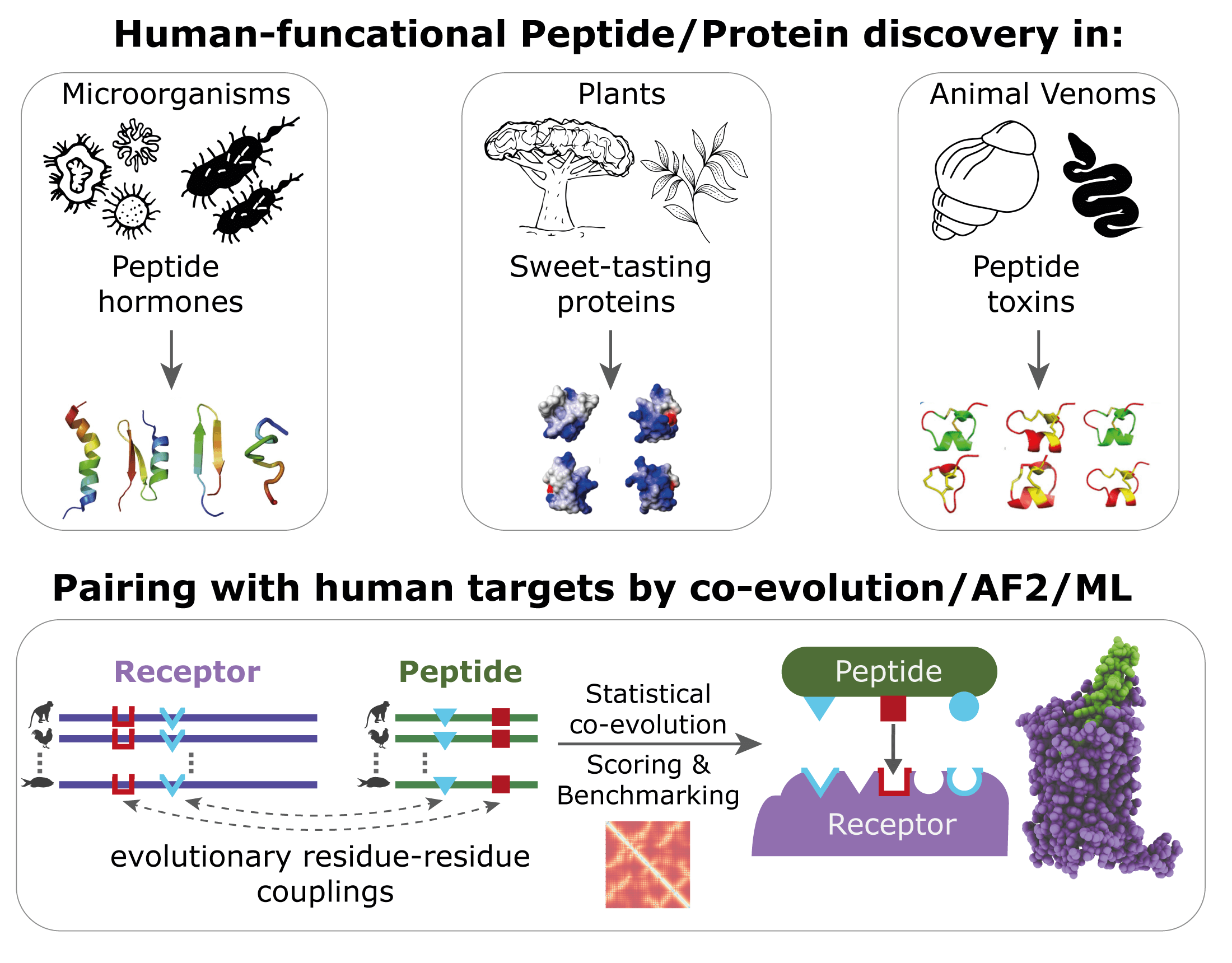 Background: Organisms constantly interact with their abiotic and biotic environment. At the molecular level, some of these interactions are driven directly or indirectly through small molecules or peptides. Peptides are genetically encoded and undergo a maturation process that may include cleavage, folding, and post translational modifications.
Background: Organisms constantly interact with their abiotic and biotic environment. At the molecular level, some of these interactions are driven directly or indirectly through small molecules or peptides. Peptides are genetically encoded and undergo a maturation process that may include cleavage, folding, and post translational modifications.
Although many bioactive peptides are produced for species-specific internal processes (e.g. fertilization or metabolisms), some are known to interact across species (e.g. predator-prey or defense) and even across domains of life (e.g. plant-human or microbial-animal).
These cross-species signals include venom-derived peptides from snails, snakes, and spiders, as well as peptides from plants, fungi, and bacteria, all of which the majority interact with membrane proteins in other organisms. The genomic revolution has allowed researchers to generate and analyse genome-scale datasets to improve our understanding of fundamental ecological and evolutionary processes.
However, two fundamental issues are critically unresolved: i) do inter-species peptide:receptor interactions at the molecular level explain ongoing ecological and evolutionary processes and ii) how do biophysical and functional relationships remain coordinated over evolutionary time.
Objective: AlphaFold2 (AF2) achieved remarkable performances on the prediction of protein structures. Moreover, there are indications that deep learning-based approaches, such as AF2, can outperform classical protein-protein docking strategies. This indicates that these strategies could be employed for the prediction of novel peptide-protein interactions, which to date, have largely been discovered through happenstance.
We are developing analytical frameworks to identify bioactive peptides across domains of life to discover and dissect novel molecular interconnections between species retelling evolutionary processes. We hypothesise that:
1. there are numerous inter-species and across domains of life peptide:receptor pairs that have co-evolved for their respective ecological niche and
2. combining genetic- and residue-level co-evolution with deep learning models will enable discovery of unknown peptide:receptor pairs indicative of new eco-evolutionary processes.
 We employ in vitro pharmacological tools to understand ligand-receptor interactions, receptor subtype selectivity, activation mechanisms, and signaling pathways at the molecular/mechanistic level.
We employ in vitro pharmacological tools to understand ligand-receptor interactions, receptor subtype selectivity, activation mechanisms, and signaling pathways at the molecular/mechanistic level.
We develop assays to characterize how human genetic variants affect the activity of approved drugs using the latest tools in GPCR pharmacology. Our in vitro experiments revealed that the effects on drug actions are vastly diverse ranging increased or reduced ligand potency and gain of unintended adverse effects elicited via alternative receptor intracellular signalling pathways.
We have ongoing projects focusing on the development of ligands with novel pharmacological profiles at G protein-coupled receptors and we seek to experimentally validate predicted endogenous and novel peptide:receptor interactions across the kingdom of life using mammalian cell-lines.
Common coupling map advances GPCR-G protein selectivity.
eLife. 2022
GPCR activation mechanisms across classes and macro/microscales.
Nature Structural & Molecular Biology. 2022
Molecular and in vivo phenotyping of genetic variants of the human glucagon receptor.
Journal of Biological Chemistry
Ligand-directed bias of G protein signaling at the dopamine D2 receptor.
Cell Chemical Biology. 2021
Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors.
Cell. 2019
Pharmacogenomics of GPCR drug targets.
Cell. 2018
Trends in GPCR drug discovery: new agents, targets and indications.
Nature Reviews Drug Discovery. 2017
Selectivity determinants of GPCR–G-protein binding.
Nature. 2017
2023 November - Independent Research Fund Denmark: Interview with Research Leader Alexander Sebastian Hauser
2023 September 27th — ISBUC article: Introducing Alexander Hauser
2021 December 2nd — SciLogs Spektrum Article: Blog post in German
2021 May 29th — Interview at Dr.GPCR: Podcast video
2021 December 2nd — Alexander S. Hauser receives Bachem Award for Peptide Science
2019 October 31st — Discovery of new hormone-receptor signaling systems: Press release
2021 March 26th — Alexander S. Hauser receives Bayer Pharmaceuticals PhD Award: Press release
2019 August 14th — Alexander Hauser received HC Ørsted research talent prize: See all prize winners here
2017 December 12th — Press release: GPCR pharmacogenomics
PhD Students/Postdocs
We are always interested in hearing from highly motivated candidates in both computational biology and/or experimental biology. Please send us a letter of intent outlining your motivation to join, as well as your publication list and CV.
Potential candidates who are interested in joining should be competitive to apply for external fellowships and sources of funding such as EMBO Long-Term Fellowship, Marie-Curie Fellowship, HFSP Fellowship or these application options for postdocs.
When available, fully funded postdoctoral fellowships will be posted here Please apply using the online system, as e-mailed applications will not be considered.
MSc/Exchange/Scholarship students
We are happy to receive inquiries from computational/bioinformatics students and/or students interested in in vitro pharmacology projects. Master students who are interested in laboratory-based projects for their final MSc thesis are typically staying for 6 - 12 months.
Group members
| Name | Title | Phone | |
|---|---|---|---|
| Harpsøe, Kasper | Research Consultant | +4535320882 | |
| Hauser, Alexander Sebastian | Associate Professor | ||
| Hoegen Dijkhof, Luuk Robin | PhD Fellow | +4535334719 | |
| Madsen, Jakob Sture | PhD Fellow | ||
| Nguyen, Trinh Trung Duong | Assistant Professor | +4535322369 | |
| Reiner-Link, David | International Researcher | +4535321328 | |
| Rönkkö, Teemu Kalle Eemeli | PhD Fellow | +4535328377 | |
| Trapkov, Boris | PhD Fellow |
| Name | Title | |
|---|---|---|
| Javier Sánchez Lorente | Master student | javier.sanchez.lorente@sund.ku.dk |
| Liam Lenihan | Erasmus student | smf595@alumni.ku.dk |
| Lucy Byrne | Erasmus student | dfc636@alumni.ku.dk |

