Gajhede Lab
The group focuses on the structural biology of selected histone demethylases and on their interactions with other enzymes involved in epigenetic regulation and with potential drug molecules. Techniques applied include recombinant protein expression in insect and mammalian cells, drug/target affinity determination using biophysical and biochemical assays, X-ray crystallography, Small Angle X-ray Scattering, Electron Microscopy, structure-based drug design and computational chemistry.
The control mechanisms for reading and translating the information stored in our DNA are referred to as epigenetics. Perhaps not surprisingly, several genes crucial in epigenetic control are also deregulated in cancer and diseases of the central nervous system. This makes the corresponding proteins very important targets for therapy. In recent years, several proteins and mechanisms that are essential for epigenetic control have been identified; one class is the histone demethylases.
Histone lysine methylation and demethylation are central events in epigenetic regulation, as they modulate the accessibility of the DNA. The major methylation sites are within histone tails at the basic amino acid side chains of lysine and arginine residues. In 2006, a member of a new class of histone demethylases, Jumonji C-terminal (JmjC) domain containing histone demethylases, was described (Yamane et al., 2006). Subsequent reports revealed that these enzymes form a large and evolutionarily conserved histone demethylase family. It has lately also been possible to map relationships between methylation and functional responses as shown in the figure below.
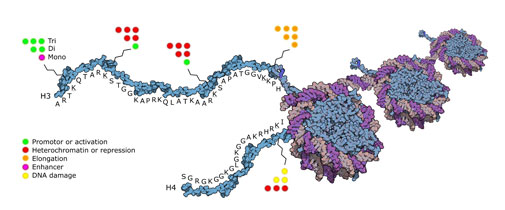
Picture above: Relation between functional responses and lysine modification
Gajhede Lab members
| Name | Title | Phone |
|---|
| Name | Title | Phone | |
|---|---|---|---|
| Narayanan, Dilip | Facility manager | ||
| Qi, Shengxiao | PhD student |