Bach Group
The PPI targets we focus on are involved in redox signalling, oxidative stress, and inflammation, making them potential drug targets for various diseases, such as metabolic dysfunction-associated steatohepatitis (MASH), chronic kidney disease (CKD), neurodegenerative disorders, and cancers.
Our main target is Keap1, which regulates Nrf2 and thereby endogenous antioxidant and anti-inflammatory responses. Additionally, we have developed inhibitors of the superoxide-generating multi-subunit enzyme complex NADPH oxidase 2 (NOX2), and recently, we expanded our focus to include tumor necrosis factor (TNF) receptors, their downstream PPIs, and novel cancer-related PPI targets.
These PPIs are challenging to target with small molecules due to their relatively large, shallow, and polar interfaces. Current inhibitors often lack affinity, cellular potency, or drug-like properties. Additionally, the proteins are involved in complex networks of intracellular interactions. Therefore, more precise and effective molecules are needed as research tools to better understand the roles of these PPIs and to exploit them as drug targets for severe diseases.
- Inhibiting the Keap1-Nrf2 PPI leads to Nrf2 activation and expression of antioxidant and anti-inflammatory proteins, thus Keap1 is a promising drug target for diseases involving oxidative stress and inflammation. Recent medicinal chemistry efforts from both academic and industrial laboratories have led to a rise in noncovalent Keap1-Nrf2 inhibitors. In our Drug Discovery Today review (Barreca and Qin et al, 2023), we provide a comprehensive overview of current noncovalent Keap1-Nrf2 inhibitors with a focus on their pharmacological effects, to examine the therapeutic potential of this promising potential future drug class. We also discuss the differences in pharmacodynamic effects and off-target profiles of covalent vs noncovalent small molecules that target Keap1 and activate Nrf2.
Further description of our original contributions to the field of Keap1 drug discovery and other targets are seen below:
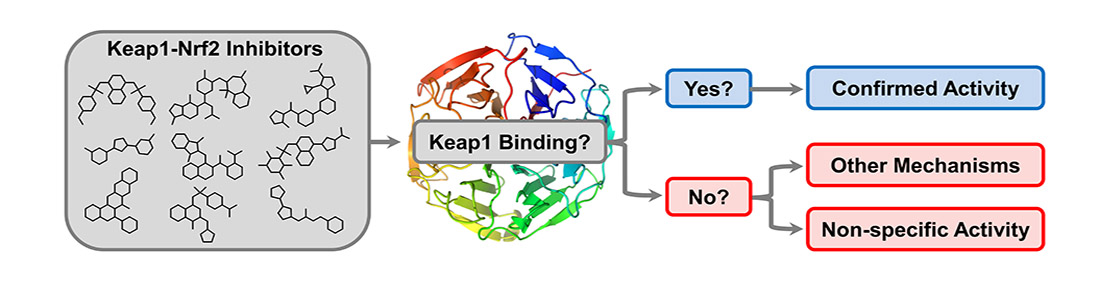
- For Keap1, we analysed known inhibitors for their ability to cross the blood-brain barrier (Pallesen et al, 2018, J Med Chem). This was followed by a comparative assessment study, where the compounds from literature were tested in a range of assays to validate and compare their activity towards Keap1. Interestingly, about half of the reported Keap1 inhibitors were found to be false positives (Tran and Pallesen et al, 2019, J Med Chem and Derek Lowe’s blog).
- We have addressed Keap1 by FBDD pursuing three strategies: First, we deconstructed the known compounds into a target-biased library of 77 fragments and tested them for Keap1 binding using four orthogonal biophysical assays. The binding modes of key fragment hits where determined by X-ray crystallography, which allowed us to merge two fragments into novel compounds with high affinities to Keap1 (Pallesen et al, 2021, J Med Chem; see also Practical Fragments blog).
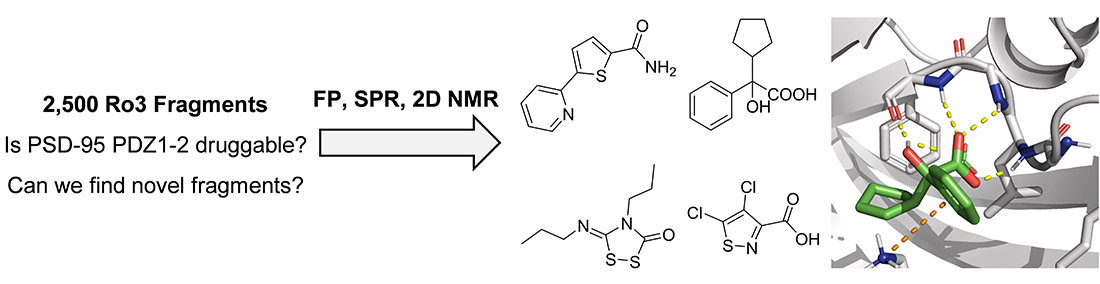
Secondly, we screened our commercial library of 2,500 fragments using four orthogonal methods. X-ray crystallography and fragment-to-lead (F2L) optimization resulted in novel fluorenone-based Keap1-Nrf2 inhibitors with a 1700-fold affinity-increase relative to the fragment hit (Narayanan and Tran et al, 2022, J Med Chem; and MAX IV’s news article).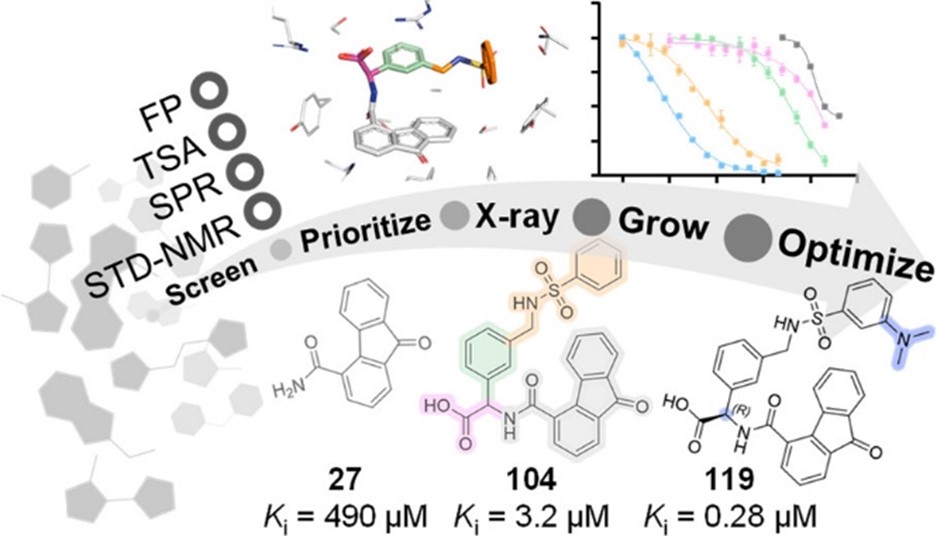
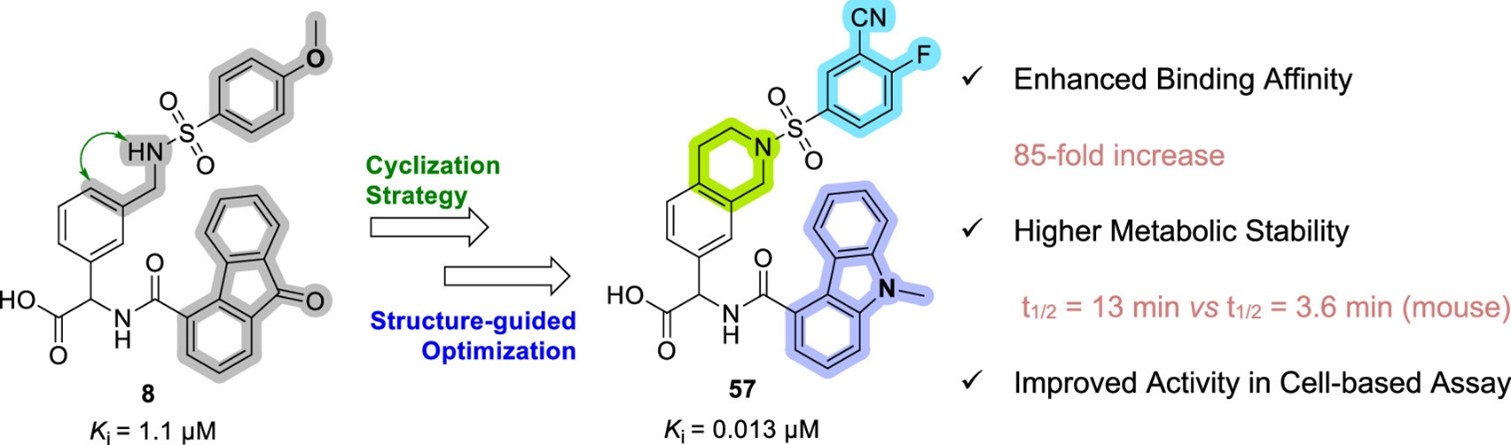
We then applied a conformational restriction strategy guided by X-ray crystallography, which led to new tetrahydroisoquinoline (THIQ) Keap1-Nrf2 inhibitors with significantly enhanced binding affinity and improved membrane permeability. Further optimization increased metabolic stability and cellular activity, and the compounds showed selectivity across a panel of homologues proteins (Qin et al, 2024, J Med Chem).
Thirdly, we have screened the Diamond Light Source’s XChem library by X-ray crystallography. F2L optimization resulted in low nanomolar affinities and cellular potent, selective, anti-inflammatory drug-like molecules. RNA sequencing revealed activation of cytoprotective pathways and a different profile from typical covalent Nrf2 activators (Lin et al, 2025, Angew. Chem. Ind. Ed.; see also Practical Fragments blog). The compounds have been patented and we are currently wrapping-up the lead optimization focusing on pharmacokinetic/pharmacodynamic properties and obtaining in vivo proof-of-concept data in models of kidney and liver diseases. We are open to partnerships and pursuing commercial opportunities.”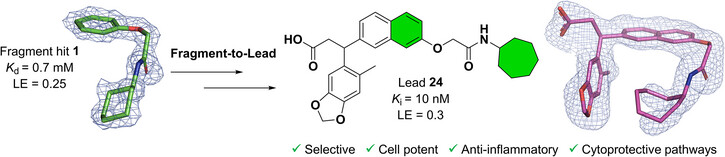
-
We have used FBDD to develop novel inhibitors of the p47phox subunit of NOX2. We screened our 2,500 fragments by fluorescence polarization (FP) and thermal shift assay (TSA) followed by surface plasmon resonance (SPR) validation. Biostructural studies by NMR and SAXS indicated that two fragments bound to two separate binding sites in the elongated conformation of p47phox, and the design of bivalent inhibitors thereby resulted in a 20-fold enhancement in affinity (Solbak et al, 2020, J Med Chem).
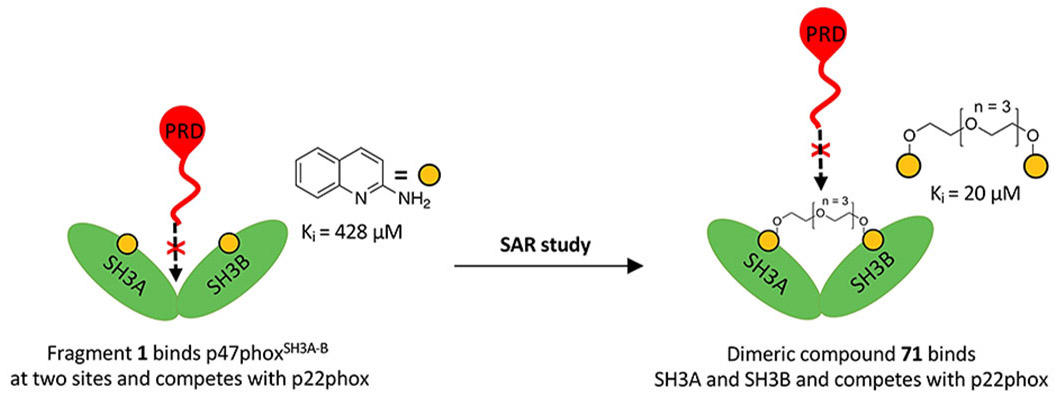
The bivalent inhibitors were then further optimized into a novel series of potent (submicromolar) NOX2 inhibitors, which were thoroughly characterized for binding to p47phox using biophysical methods and cellular activities in different cell lines. With this, we showed that p47phox can be targeted by potent small molecules, which may inspire future development of chemical probes and drug leads (Zang et al, 2023, J Med Chem). We have also assessed a literature compound suggested to target p47phox and inhibit NOX2. We found that the compound, LMH001, degraded within minutes in buffer and did not inhibit the p47phox/p22phox interaction, as suspected (Zang et al, 2023, Front Pharmacol).
- PDZ domains are intriguing but challenging drug targets. We have investigated the druggability of PSD-95’s PDZ1-2 domain by fragment-based screening and a computational method. This resulted in new fragments shown to bind the PDZ domains of PSD-95 (Zang et al, 2020, ChemMedChem).
- We are engaged in many collaboration projects: We used SPR to characterize protein-protein interactions related to KDM5B (Dorosz et al, 2019, Sci Rep), NEMO (Jussupow et al, 2020, Sci Adv), and CaMKIIa (Leurs et al, 2021, PNAS USA). We contributed with LC-MS pharmacokinetic data to a small-molecule obesity study (Grunddal et al, 2021, Mol Metab); and, we designed and synthesized novel Cas9 inhibitors by using a fragment-inspired deconstruction-reconstruction optimization strategy of a screening hit (Lee and Tran et al, 2022, J Med Chem).
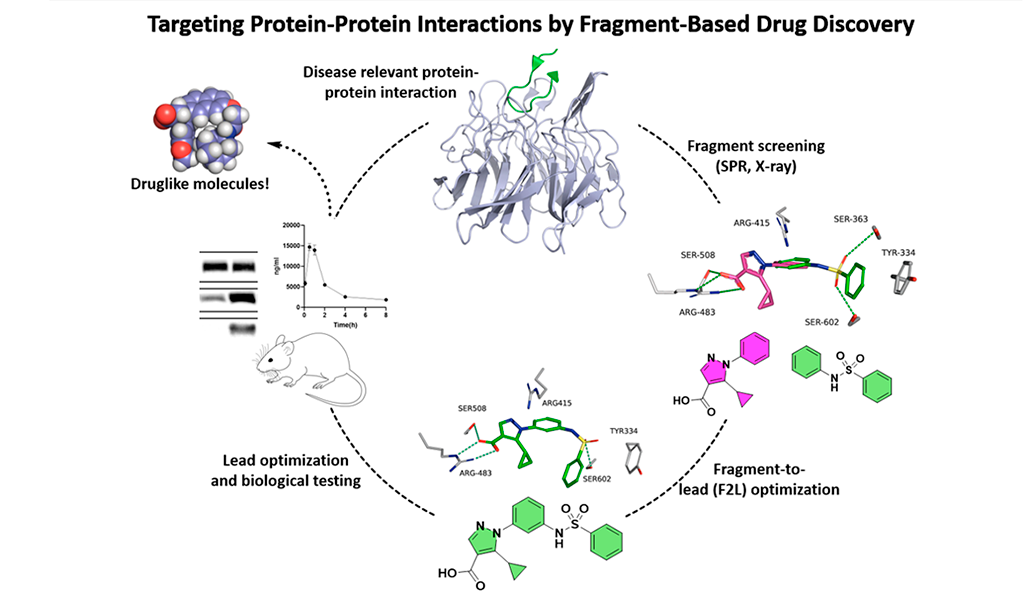
We screen our libraries of fragments (small substructures of drug-like molecules) by sensitive biophysical methods (FP, TSA, SPR, ligand-based NMR). Promising hits are optimized into lead compounds by medicinal chemistry, X-ray crystallography, and pharmacological assays.
Medicinal chemistry, SPR (see SelectScience interview about our Pioneer system), and other biochemical/biophysical assays are core methods of our group. X-ray crystallography is performed in close collaboration with internal colleagues Dr. Dilip Narayanan and Prof. Michael Gajhede using various beamlines in Europe, e.g. ESRF (Grenoble), EMBL (Hamburg), MAX IV (Lund), and Diamond Light Source (Oxfordshire). NMR for studying ligand-protein interactions has been done with collaborators in Germany (Helmholtz Zentrum München) and Denmark (Technical University of Denmark), and testing in cell assays and disease models by collaborators in Denmark (University of Copenhagen, University of Southern Denmark, Aarhus University), Sweden (Karolinska Institute), and USA (University of Miami and Fort Lewis College).
We are part of UBIMOTIF – a Marie Sklodowska-Curie Innovative Training Network (MSC-ITN) – focusing on the ubiquitin system and E3 ligases; the COST Action ‘CA20121 - Bench to bedside transition for pharmacological regulation of NRF2 in noncommunicable diseases (BenBedPhar)’; and a collaborative project on TNF receptors with Prof. Mads H. Clausen from the Technical University of Denmark (see our review here).
Assistant Professors
- Dilip Narayanan (Sept 2021 - March 2024)
Postdocs
- Jie Zang (Feb 2021 – Feb 2024)
- Chunyu Lin (Feb 2023 – March 2024)
- Dilip Narayanan (June 2016 – March 2020)
- Jakob Staun Pallesen (July – Nov 2019)
- Sara Marie Øie Solbak (March 2016 – March 2019)
PhD students
- Yuting Qin (Nov 2020 – April 2024)
- Chunyu Lin (Nov 2019 - Dec 2022)
- Jie Zang (Nov 2017 – Feb 2021)
- Jakob Staun Pallesen (Jan 2016 – June 2019)
Guest Researchers
- Marilia Barreca (Postdoc) (June 2022 – Jan 2023)
- Giuseppe Marseglia (PhD Student) (Nov 2018 – April 2019)
Research assistants
- Kim Tai Tran (May 2019 – Aug 2020)
- Lars Jakobsen Høj (Sept – Dec 2017)
MSc students
- Livia Guiggi (Erasmus) (Jan 2024 – July 2024)
- Jingyi Wang (Sept 2023 – June 2024)
- Katrine Povlsen (incl. Scholar project) (Sept 2022 – March 2024)
- Lars Henrik Svensson (Sept 2022 – Jan 2023)
- Niels Guldager (Nov 2021 – July 2022)
- Frederik Wong Christensen (Nov 2020 – Aug 2021)
- Felix Peters (Erasmus) (March 2020 – Feb 2021)
- Louis Martin Eichstedt Sørensen (Nov 2019 – Dec 2020)
- Munira Mohamed Shishay Hissabu (Feb – Nov 2020)
- Kristina Olegovna Vasilyeva (Feb – Aug 2020)
- Elina Mukminova (Sept 2019 – Aug 2020)
- Martin Mariboe Olesen (Sept 2019 – Aug 2020)
- Amina Baig (Sept 2018 – March 2020)
- Dorleta Gonzalez Chichon (Sept 2018 – Aug 2019)
- Erik Bjørn Dampe (Sept 2018 – Aug 2019)
- Martina Luchini (Erasmus) (March – Aug 2019)
- Kim Tai Tran (incl. Scholar project) (Sept 2017 – April 2019)
- Nanna Haapanen (Sept 2017 – Jan 2019)
- Rosa Macarena Carrasquilla Carmona (Sept 2016 – Aug 2017)
- Lars Jakobsen Høj (Feb – Aug 2017)
- Anthony Garcia (Erasmus) (Feb – Aug 2017)
- Federico Munafo (Erasmus) (Oct 2016 – March 2017)
- Alejandro Aguayo Orozco (Sept 2014 – Aug 2015)
- Thomas Breum Pedersen (Sept 2014 – Aug 2015)
Other students
- Nikolaj Holst-Andersen (BSc) (Feb – June 2024)
- Dimitra Vlissari (Individualized study unit) (Feb – June 2024)
- Rita Turcio (Erasmus exchange) (April 2023 – Sept 2023)
- Helene Kirstine Balslev (BSc) (Feb – June 2023)
- Simon Strange Wismann (Self-funded scholarship) (Aug 2022 – June 2023)
- Georgia Goutsiou (Erasmus exchange ) (March – Oct 2020)
- Alejandro Escobar Peso (Erasmus exchange) (Oct 2016 – Jan 2017)
- Kim Tai Tran (BSc) (Jan – June 2016)
We thank the following foundations for their generous support of our research:
⦁ Independent Research Fund Denmark (Project 1)
⦁ SPARK Denmark (Novo Nordisk Foundation)
⦁ Innovation Fund Denmark
⦁ The Carlsberg Foundation
⦁ Lundbeck Foundation (Fellowship and Ascending investigator grants)
⦁ Independent Research Fund Denmark (Project 2 grant with Prof. Clausen)
⦁ MSC-ITN (UBIMOTIF) (PhD stipend with Prof. Jakob Nilsson)
⦁ BioInnovation Institute (BII) (Proof of concept grant)
⦁ China Scholarship Council
⦁ HALOS Cross Border Research Grant
⦁ Torben and Alice Frimodt´s Foundation
⦁ Simon Spies Foundation
⦁ Hørslev Foundation
⦁ Augustinus Foundation
If you are you interested in doing a master thesis project in the group, please send your CV, grades and motivations to anders.bach@sund.ku.dk for further discussions. We have projects both within chemistry and biological areas of drug discovery.
- Example of a Master Thesis Project:
Fragment-based drug discovery on protein-protein interactions (pdf)
Currently we have no open PhD and postdoc positions in our group, but we are always interested in hearing from talented and potential candidates. If you are interested in becoming part of our group please send your CV and motivations to anders.bach@sund.ku.dk
Group members
| Name | Title | Phone | |
|---|---|---|---|
| Anders Bach | Professor | +4535336242 | |
| Camilla Bachmand Chan | Guest Researcher | ||
| Fabiana Lo Mascolo | Visiting PhD Student | ||
| Katrine Povlsen | PhD Fellow | ||
| Marie Elodie Hélène Cadot | Postdoc | +4535333819 | |
| Rebecca Storm Böttern Olesen | Master Thesis Student | ||
| Richard Nekanovic | Master Thesis Student | ||
| Salomi Georgiou | Master Thesis Student | ||
| Thomas Svava Mortensen | Research Assistant |