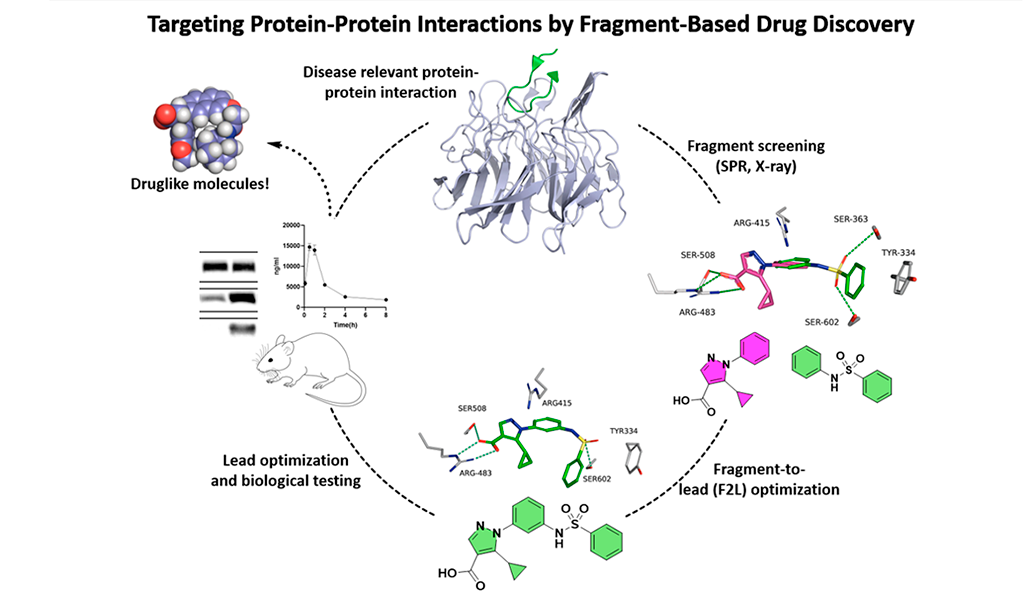
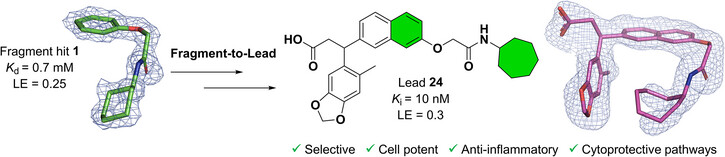
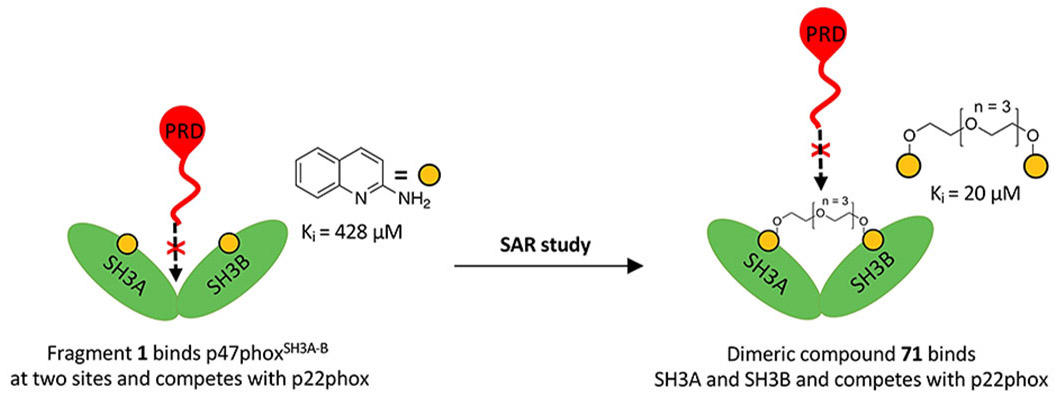
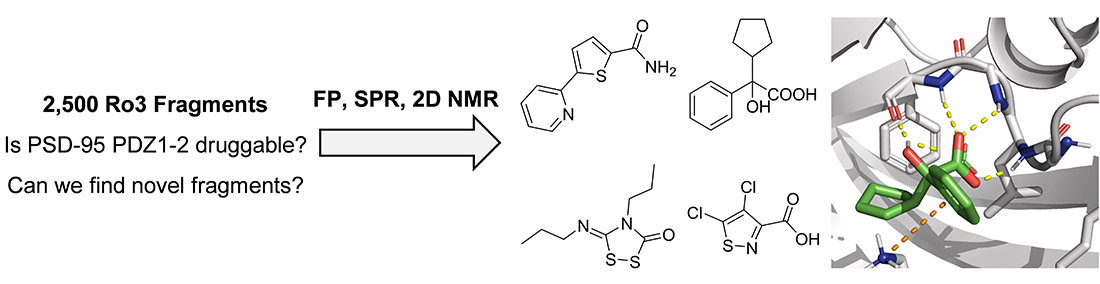
The PPI targets we focus on are involved in redox signalling, oxidative stress, and inflammation, making them potential drug targets for various diseases, such as metabolic dysfunction-associated steatohepatitis (MASH), chronic kidney disease (CKD), neurodegenerative disorders, and cancers.
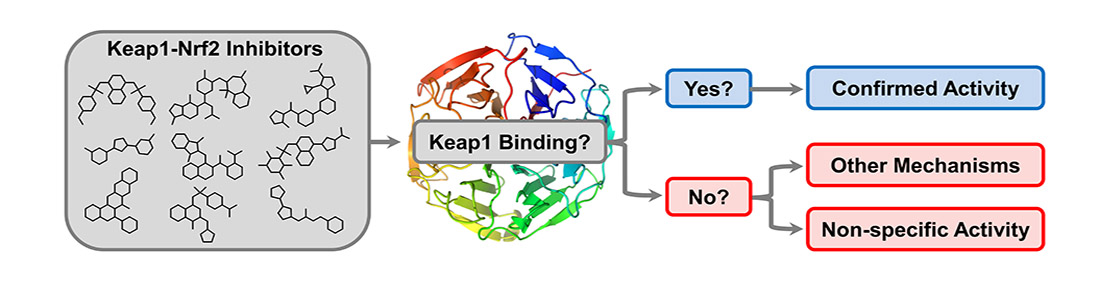
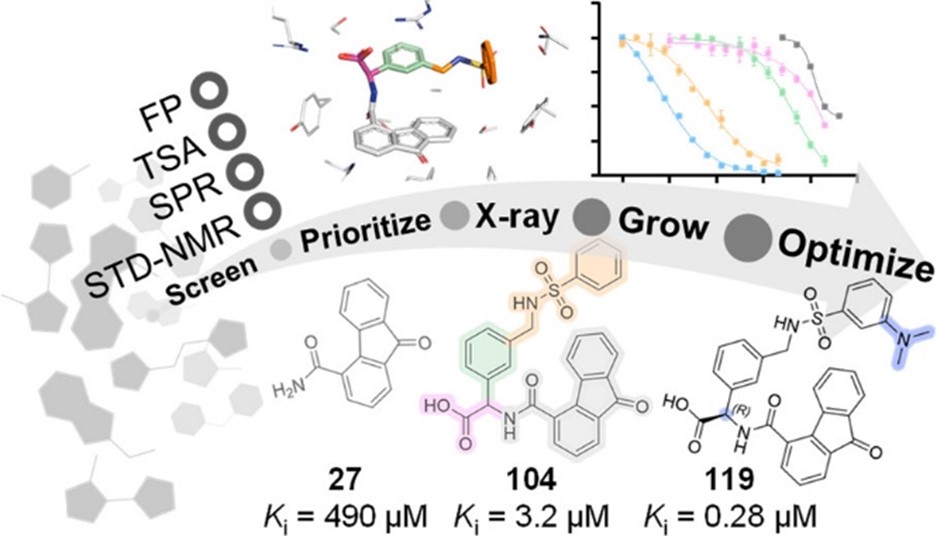
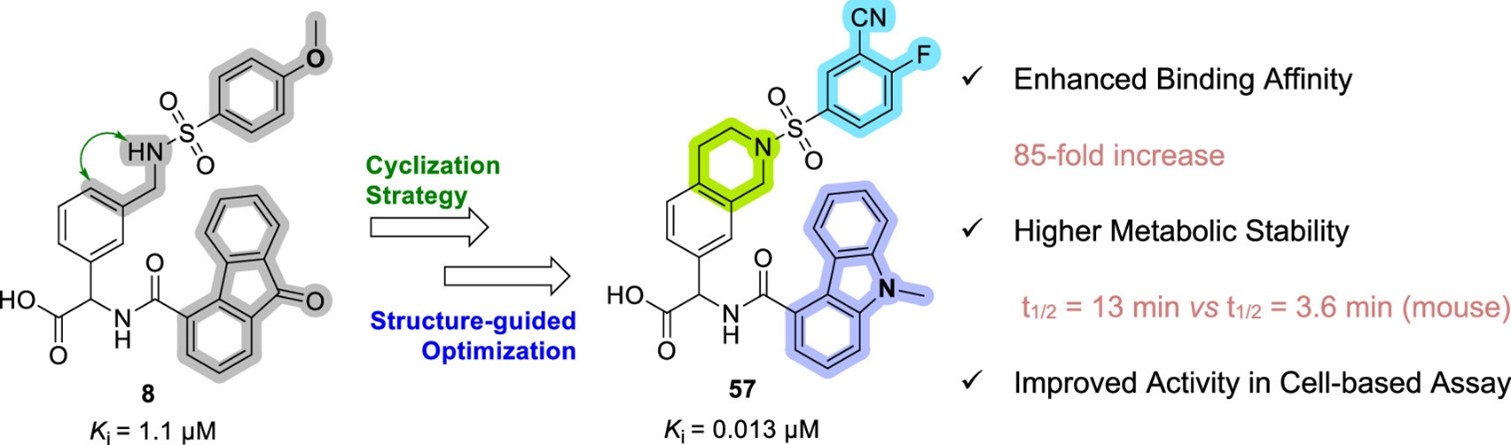
Our main target is Keap1, which regulates Nrf2 and thereby endogenous antioxidant and anti-inflammatory responses. Additionally, we have developed inhibitors of the superoxide-generating multi-subunit enzyme complex NADPH oxidase 2 (NOX2), and recently, we expanded our focus to include tumor necrosis factor (TNF) receptors, their downstream PPIs, and novel cancer-related PPI targets.
These PPIs are challenging to target with small molecules due to their relatively large, shallow, and polar interfaces. Current inhibitors often lack affinity, cellular potency, or drug-like properties. Additionally, the proteins are involved in complex networks of intracellular interactions. Therefore, more precise and effective molecules are needed as research tools to better understand the roles of these PPIs and to exploit them as drug targets for severe diseases.